Platform
Expanding the drug design paradigm
Cullgen’s ubiquitin-mediated, small molecule-induced target elimination technology, uSMITE™ is at the forefront to develop novel therapeutics for individuals with various difficult to treat diseases, including inflammation, autoimmune diseases, neurodegenerative disease and cancer.
Traditionally, small molecule drugs are designed to interact with the functional sites of proteins and block their activities. We are developing uSMITE™ to expand the drug design paradigm beyond functional site inhibition, to make it possible to eliminate previously “undruggable” enzymes and proteins by targeted destruction. We also intend to use the uSMITE™ technology to harness the ubiquitin proteasome system, a multi-step biochemical process that controls protein degradation in all cells. From years of research on the ubiquitin-proteasome system and key discoveries about E3 ubiquitin ligases, Cullgen’s founders have already demonstrated that the underlying technology can rapidly generate a large number of highly potent, selective, and bioavailable compounds targeting various disease-causing proteins. Furthermore, this process is significantly more cost effective when compared to traditional drug discovery approaches.
The ubiquitin-proteasome system
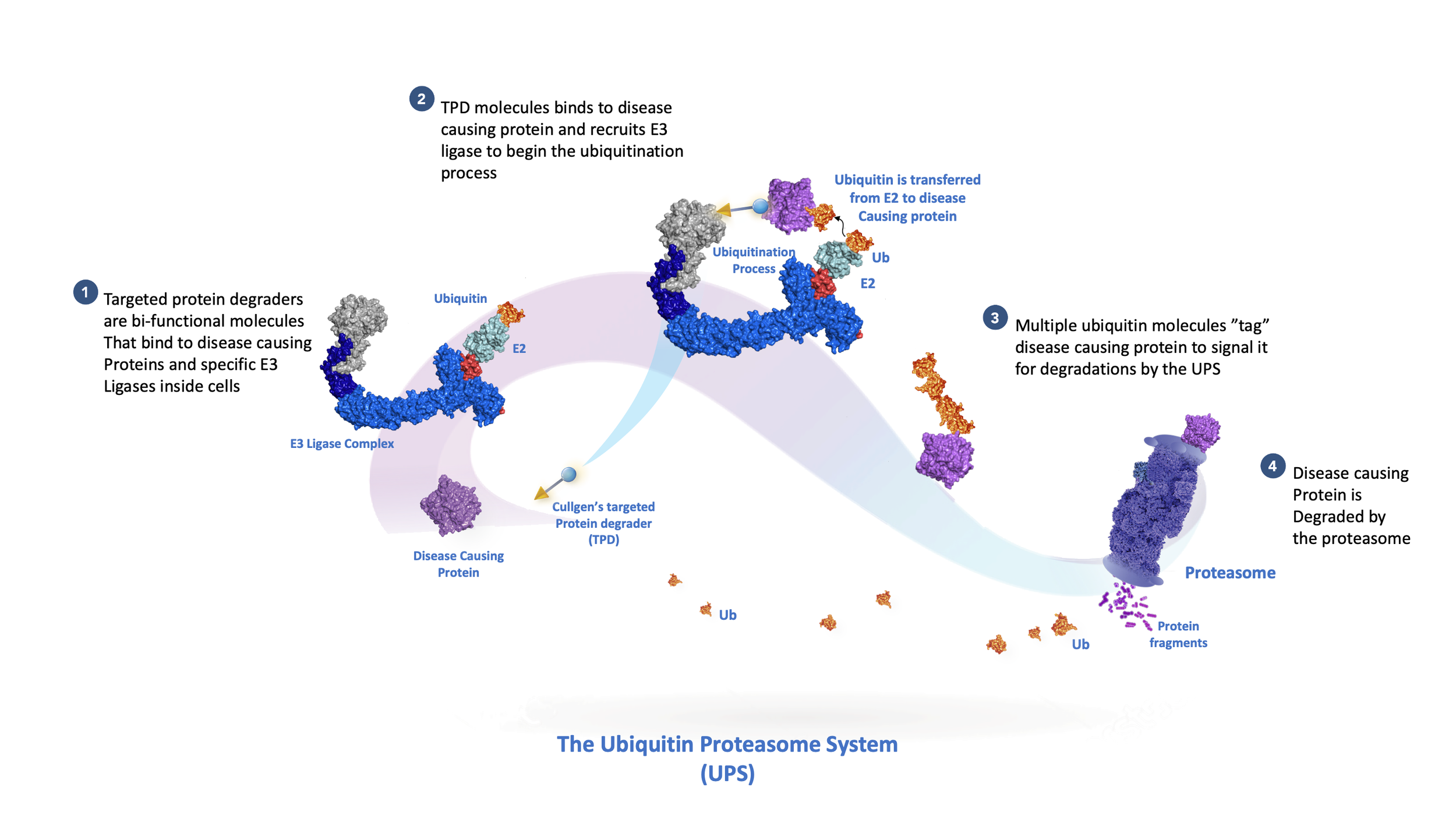
Ubiquitin is a small (76 amino acid residues) protein that is present in all organisms and, as the name suggests, is expressed ubiquitously in all cells in our body. Ubiquitin is joined to other proteins (substrates) by a process known as ubiquitylation. Ubiquitylation can conjugate either a single ubiquitin (monoubiquitylation) or create a chain of ubiquitins (polyubiquitylation) on lysine residues of substrate proteins. The polyubiquitin chain functions as an ID tag that marks substrate proteins for the delivery to the proteasome, a large protein complex in which protein degradation takes place.
All proteins, sooner or later, are degraded. Degradation is one means to regulate the amounts or activities of proteins in a cell. The ubiquitin-proteasome system (UPS) regulates virtually all cellular processes, such as cell growth, proliferation, migration, signaling, and death. Thus, impairments in the UPS cause many human diseases, including developmental retardation, neurodegenerative diseases, immune diseases, heart diseases, and cancer. The importance of the UPS was underscored by awarding of the 2004 Nobel Prize in Chemistry to Aaron Ciechanover, Avram Hershko, and Irwin Rose, who were instrumental in elucidating the biochemical details of the ubiquitylation.
Video 1 - The UPS System
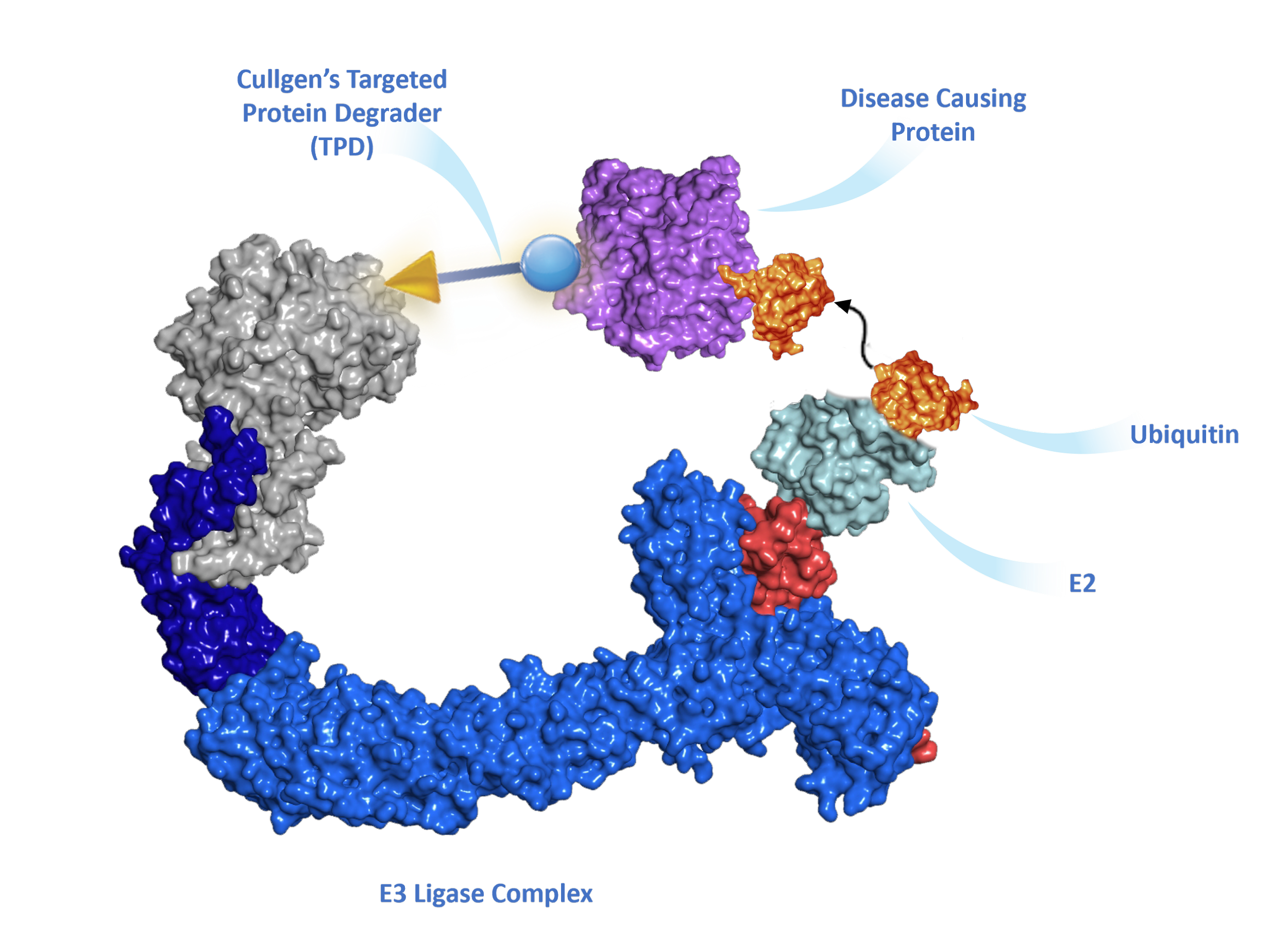
Tagging substrate proteins with ubiquitin, or ubiquitylation, is a highly sophisticated biochemical process catalyzed by a cascade of three enzymes. First, ubiquitin is “activated” by being transferred to a ubiquitin-activating enzyme (E1); this reaction requires the energy-rich molecule ATP. Second, the activated ubiquitin is transferred from E1 to a ubiquitin-conjugating enzyme (E2). Finally, a ubiquitin ligase (E3) transfers the ubiquitin from E2 to a lysine residue in the substrate protein. The E3 ligase determines when, where, and which proteins are tagged with ubiquitin. Human cells possess an estimated more than 600 E3 ligases, each specific for particular protein targets. Building upon this precise degradation system and 20 years of research experiences, scientists at Cullgen are exploring novel strategies to target selective disease-causing proteins for the degradation by the UPS.
E3 ligases can be used by small molecules to degrade disease proteins
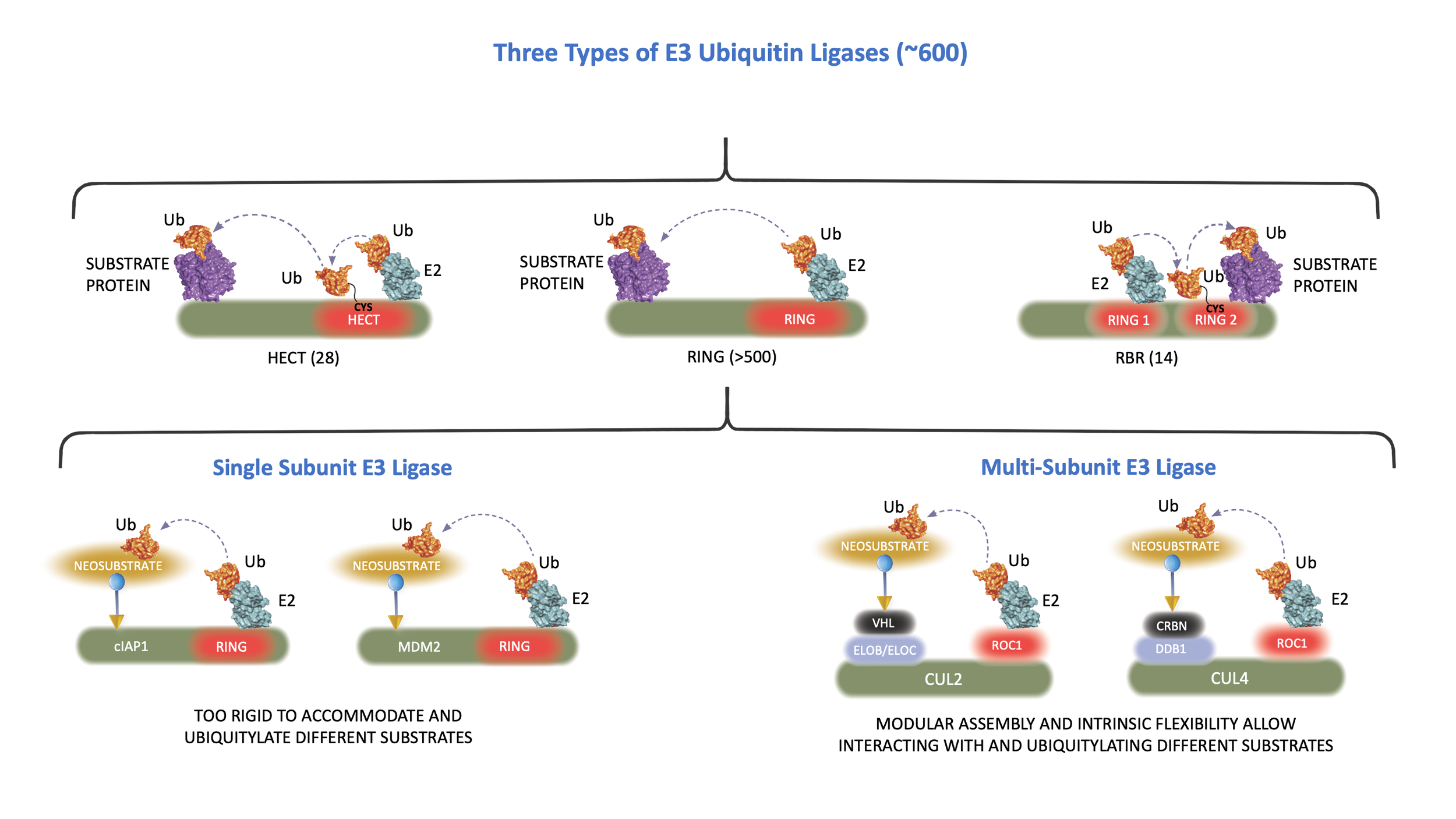
The vast majority of E3 ubiquitin ligases have a structure known as the RING finger. The RING finger mediates ubiquitin transfer from E2 to a target protein. There are two major types of RING finger E ligases: One containing an intrinsic RING finger domain and the other using a scaffold protein, cullin, to bind to a small RING finger protein, ROC1 or ROC2. Individual cullin proteins can bind with multiple different substrate receptor proteins, either directly, such as CUL3, or indirectly through a linker protein, such as ELOB/ELOC in the case of cullin 2 (CUL2) and DDB1 in the case of CUL4. Each cullin can associate with many different substrate receptors, leading to the assembly of an estimated more than 300 distinct cullin-RING E3 ubiquitin ligases (CRLs). The modular assembly of CRL ligase complexes provides the flexibility to specifically target the degradation of a large number of substrate proteins. This flexibility can also be reprogrammed by small molecules to degrade proteins of interest.
Recognizing its potential, scientists at Cullgen are developing small molecules to target the degradation of disease-causing proteins. Small molecule-mediated selective targeting of protein degradation represents an unprecedented opportunity in drug discovery and has several advantages over traditional drug discovery strategies. These advantages include the high degree of target specificity that can be rapidly validated, the potential to reduce systemic drug exposure, and the ability to counteract increased target protein expression induced by the inhibition of protein function. Importantly, small molecule-induced protein degradation is not limited (or even directed) to the functional sites of enzymes and can be applied to target proteins that are not currently therapeutically tractable, such as transcription factors, scaffold, and other non-enzyme regulatory proteins. The dependency of protein degradation on the E3 ligase and vast repertoire of E3 ligases makes it possible to develop novel E3 ligands to target the degradation of proteins of interest in a selective tissue, tumor type and subcellular compartment.